What is DeepNitro?
DeepNitro is a useful tool that predicts the sites of protein Nitration and S-Nitrosylation. By utilizing Deep Learning algorithm, we trained three prediction models based on the published modification data for Tyr-Nitration, Trp-Nitration and S-Nitrosylation. In DeepNitro, we tried to make it powerful and convenient to be used by combining the three models. The online service and local packages were provided. This page is prepared as the usage for the online service. For usage of local packages, please download the Manual.
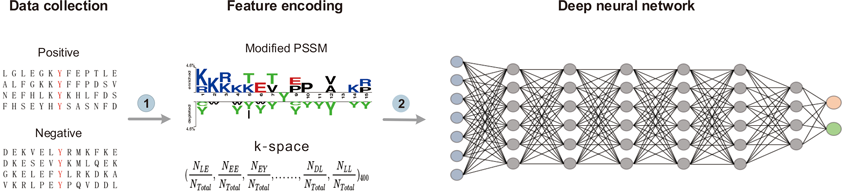
How do I use it?
1. Firstly, to use DeepNitro you will need to upload a protein data file or paste a formatted test in FASTA format.
An example for FASTA format:
>sp|P31751|AKT2_HUMAN RAC-beta serine/threonine-protein kinase OS=Homo sapiens GN=AKT2 PE=1 SV=2 MNEVSVIKEGWLHKRGEYIKTWRPRYFLLKSDGSFIGYKERPEAPDQTLPPLNNFSVAEC QLMKTERPRPNTFVIRCLQWTTVIERTFHVDSPDEREEWMRAIQMVANSLKQRAPGEDPM DYKCGSPSDSSTTEEMEVAVSKARAKVTMNDFDYLKLLGKGTFGKVILVREKATGRYYAM KILRKEVIIAKDEVAHTVTESRVLQNTRHPFLTALKYAFQTHDRLCFVMEYANGGELFFH LSRERVFTEERARFYGAEIVSALEYLHSRDVVYRDIKLENLMLDKDGHIKITDFGLCKEG ISDGATMKTFCGTPEYLAPEVLEDNDYGRAVDWWGLGVVMYEMMCGRLPFYNQDHERLFE LILMEEIRFPRTLSPEAKSLLAGLLKKDPKQRLGGGPSDAKEVMEHRFFLSINWQDVVQK KLLPPFKPQVTSEVDTRYFDDEFTAQSITITPPDRYDSLGLLELDQRTHFPQFSYSASIR E
Note: In order to guarantee a safe run of our web server, a maximal file size of 2M is allowed to be uploaded in each case.
2. Next, select the PTM types and thresholds. The PTM Type panel lists 3 options, S-Nitrosylation, Tyrosine Nitration and Tryptophan Nitration. For a better prediction performance,three thresholds of high, medium and low stringency were chosen for each PTM type.In order to balance the prediction performance, the medium stringency was selected as the default threshold.In addition, we offer several examples to try out the web server.
3. Finally, select a prediction threshold for each PTM type to start the calculation. The current status can be view in the Result panel.
4. All of the prediction results are presented in a tabular form containing the information of FASTA title ID, modified position, modified peptide, predicted score, prediction cutoff and modified type. In the position column, the precise modification sites are shown. Also, the predicted peptide for modification is displayed in the peptide column with the modified site shown in red. The cluster score and its corresponding cutoff are presented in the score and cutoff column, respectively. In the last column, the explicit PTM type is indicated.
What is the prediction threshold?
The prediction models of DeepNitro were trained using Deep Learning algorithm. After training the prediction models, we carried out 10-fold cross-validation to validate the prediction performance. To balance the prediction accuracy, we selected three thresholds with high, medium and low stringencies based on the evaluation results. The detailed performance under these three thresholds was presented as follow:
What is the DeepNitro Database?
DeepNitro Database collected the information of proteins that could be Nitrated or S-Nitrosylated. So far, 2577 S-Nitrosylated proteins, 755 tyrosine-nitrated proteins and 51 tryptophan-nitrated protein have been included in DeepNitro Database. Besides the modification data, users also can query or browse contains other protein details from DeepNitro database. Data Summary of DeepNitro database:
How to use the Database?
1. Search. You can input one or multiple keywords (separated by space character) to search the DeepNitro database. The search fields including DeepNitroDB ID, UniProt Accession, Protein Name, Protein Alias, Gene Name, Gene Alias, Species, and Function.
EXAMPLE: Please click on the "Example" button to search "kinase " in Function field. By clicking on the "Submit" button, the related proteins will be shown.
2. Advance search allows you to input up to three terms to find the information more specifically. The querying fields can be empty if less terms are needed. The three terms could be connected by the following operators:
exclude: If selected, the term following this operator must be not contained in the specified field(s)
and: the term following this operator has to be included in the specified field(s)
or: either the preceding or the following term to this operator should occur in the specified field(s)
EXAMPLE: You can click on the "Example" button to load an instance, which could search S-Nitrosylated ribosomal protein in human. The human B4DMJ2 (DB-9606-00030), which is nitrosylated at C96 will be shown by clicking on the "Submit" button.
3. Browse. You can browse the DeepNitro database by species. With the default setting, you can directly click on the "Submit" button to browse all the proteins that are modified by Nitro or SNO groups in the database.
4. BLAST search could be used to find the specific protein and/or related homologues by sequence alignment. This search-option will help you to find the querying protein accurately and fast. Only one protein sequence in FASTA format is allowed per time. The E-value threshold could be user-defined, while the default is 0.01.
EXAMPLE: You can click on the "Example" button toload the protein sequence of human Rat L-lactate dehydrogenase A chain. By clicking on the "Submit" button, you can find the related homologues in other species.
I'm having trouble with DeepNitro that isn't addressed on this page. What should I do?
If you are having trouble with DeepNitro please contact the two major authors: Dr. Jian Ren and Dr. Zhixiang Zuo. We will try to resolve it.