DeepNitro is a useful tool for prediction of protein Nitration and S-Nitrosylation
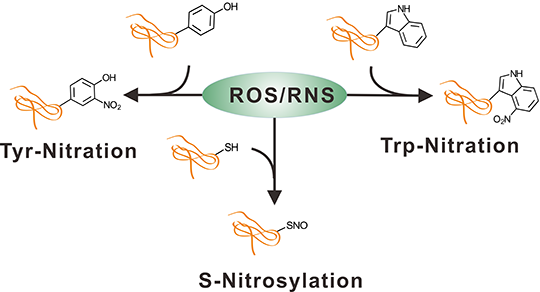
Protein Nitration and S-Nitrosylation are important post-translational modifications caused by reactive nitrogen species (RNS) compounds;
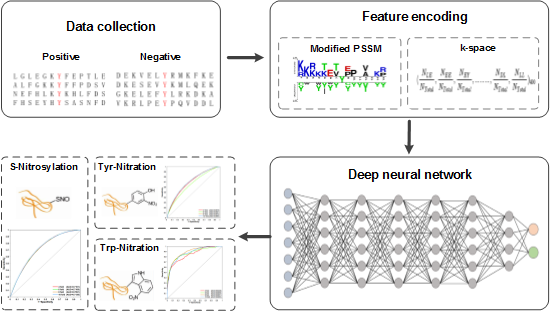
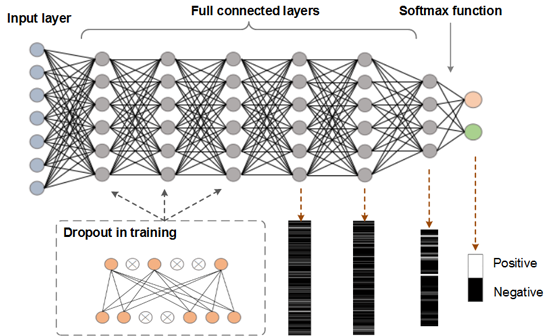
By utilizing Deep Learning, DeepNitro has developed a new algorithm for prediction of protein modification sites;
DeepNitro can simultaneously predict the exact locations of Tyr-Nitration, Trp-Nitration and S-Nitrosylation;